BC3203
Expected outputs for Tutorial 4 exercises
Question 1: Your answer should produce a tibble that looks like this;
## # A tibble: 6 × 8
## GeneID protein_name `133hpf` `29hpf` Egg t12h t2h t72h
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1.2.1045.m1 Homeobox protein … 0.0328 -1.44 -1.67 2.55 0.259 1.27
## 2 1.2.1655.m1 Homeobox protein … 0.0221 -0.204 -0.258 0.260 0.0283 0.386
## 3 1.2.20276.m1 Homeobox protein … 0.0319 -0.364 -0.340 0.359 0.0426 0.540
## 4 1.2.20289.m1 Homeobox protein … 0.0207 0.0845 -0.172 0.0567 0.0425 0.0746
## 5 1.2.21324.m1 Homeobox protein … 0.628 -0.681 -0.652 -0.0238 0.867 -0.126
## 6 1.2.25539.m1 Homeobox protein … 1.27 -1.50 -1.57 0.737 1.23 0.681
Question 2: Your answer should produce a tibble that looks like this;
## # A tibble: 6 × 8
## GeneID protein_name `133hpf` `29hpf` Egg t12h t2h t72h
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1.2.1045.m1 Homeobox protein … 0.0328 -1.44 -1.67 2.55 0.259 1.27
## 2 1.2.1655.m1 Homeobox protein … 0.0221 -0.204 -0.258 0.260 0.0283 0.386
## 3 1.2.20276.m1 Homeobox protein … 0.0319 -0.364 -0.340 0.359 0.0426 0.540
## 4 1.2.20289.m1 Homeobox protein … 0.0207 0.0845 -0.172 0.0567 0.0425 0.0746
## 5 1.2.21324.m1 Homeobox protein … 0.628 -0.681 -0.652 -0.0238 0.867 -0.126
## 6 1.2.25539.m1 Homeobox protein … 1.27 -1.50 -1.57 0.737 1.23 0.681
Question 3: Your answer should produce a tibble that looks like this;
## # A tibble: 6 × 8
## GeneID protein_name `133hpf` `29hpf` Egg t12h t2h t72h
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1.2.1045.m1 Homeobox protein … 0.0328 -1.44 -1.67 2.55 0.259 1.27
## 2 1.2.1655.m1 Homeobox protein … 0.0221 -0.204 -0.258 0.260 0.0283 0.386
## 3 1.2.20276.m1 Homeobox protein … 0.0319 -0.364 -0.340 0.359 0.0426 0.540
## 4 1.2.20289.m1 Homeobox protein … 0.0207 0.0845 -0.172 0.0567 0.0425 0.0746
## 5 1.2.21324.m1 Homeobox protein … 0.628 -0.681 -0.652 -0.0238 0.867 -0.126
## 6 1.2.25539.m1 Homeobox protein … 1.27 -1.50 -1.57 0.737 1.23 0.681
Question 4: Your answer should produce a tibble that looks like this;
## # A tibble: 6 × 8
## GeneID protein_name `133hpf` `29hpf` Egg t12h t2h t72h
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1.2.1045.m1 Homeobox protein… 0.0328 -1.44 -1.67 2.55 0.259 1.27
## 2 1.2.1655.m1 Homeobox protein… 0.0221 -0.204 -0.258 0.260 NA 0.386
## 3 1.2.20276.m1 Homeobox protein… 0.0319 -0.364 -0.340 0.359 0.0426 0.540
## 4 1.2.20289.m1 Homeobox protein… 0.0207 0.0845 -0.172 0.0567 0.0425 0.0746
## 5 1.2.21324.m1 Homeobox protein… 0.628 -0.681 -0.652 -0.0238 0.867 -0.126
## 6 1.2.25539.m1 Homeobox protein… 1.27 -1.50 -1.57 0.737 1.23 0.681
Question 5: Your answer should produce a tibble that looks like this;
## # A tibble: 6 × 6
## `133hpf` `29hpf` Egg t12h t2h t72h
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.0328 -1.44 -1.67 2.55 0.259 1.27
## 2 0.0221 -0.204 -0.258 0.260 0.0283 0.386
## 3 0.0319 -0.364 -0.340 0.359 0.0426 0.540
## 4 0.0207 0.0845 -0.172 0.0567 0.0425 0.0746
## 5 0.628 -0.681 -0.652 -0.0238 0.867 -0.126
## 6 1.27 -1.50 -1.57 0.737 1.23 0.681
Question 6: Your answer should produce a named vector that looks like this;
## 133hpf 29hpf Egg t12h t2h t72h
## 0.3346572 -0.6842727 -0.7779914 0.6567760 0.4109053 0.4708123
Wrangling
Question 7: Your answer should produce a tibble that looks like this;
## # A tibble: 36 × 4
## GeneID protein_name Stage Expression
## <chr> <chr> <chr> <dbl>
## 1 1.2.1045.m1 Homeobox protein NK-2 homolog E 133hpf 0.0328
## 2 1.2.1045.m1 Homeobox protein NK-2 homolog E 29hpf -1.44
## 3 1.2.1045.m1 Homeobox protein NK-2 homolog E Egg -1.67
## 4 1.2.1045.m1 Homeobox protein NK-2 homolog E t12h 2.55
## 5 1.2.1045.m1 Homeobox protein NK-2 homolog E t2h 0.259
## 6 1.2.1045.m1 Homeobox protein NK-2 homolog E t72h 1.27
## 7 1.2.1655.m1 Homeobox protein HLX1 133hpf 0.0221
## 8 1.2.1655.m1 Homeobox protein HLX1 29hpf -0.204
## 9 1.2.1655.m1 Homeobox protein HLX1 Egg -0.258
## 10 1.2.1655.m1 Homeobox protein HLX1 t12h 0.260
## # ℹ 26 more rows
Question 8: Your answer should produce a tibble that looks like this;
## # A tibble: 36 × 4
## GeneID protein_name Stage Expression
## <chr> <chr> <chr> <dbl>
## 1 1.2.1045.m1 Homeobox protein NK-2 homolog E 133hpf 0.0328
## 2 1.2.1655.m1 Homeobox protein HLX1 133hpf 0.0221
## 3 1.2.20276.m1 Homeobox protein NK-2 homolog B 133hpf 0.0319
## 4 1.2.20289.m1 Homeobox protein Hox-7 133hpf 0.0207
## 5 1.2.21324.m1 Homeobox protein Uncx4.1 133hpf 0.628
## 6 1.2.25539.m1 Homeobox protein six1a 133hpf 1.27
## 7 1.2.1045.m1 Homeobox protein NK-2 homolog E 29hpf -1.44
## 8 1.2.1655.m1 Homeobox protein HLX1 29hpf -0.204
## 9 1.2.20276.m1 Homeobox protein NK-2 homolog B 29hpf -0.364
## 10 1.2.20289.m1 Homeobox protein Hox-7 29hpf 0.0845
## # ℹ 26 more rows
Question 9: Your answer should produce a tibble that looks like this;
## # A tibble: 12 × 3
## GeneID Stage Expression
## <chr> <chr> <dbl>
## 1 1.2.1045.m1 133hpf 0.0328
## 2 1.2.1655.m1 133hpf 0.0221
## 3 1.2.20276.m1 133hpf 0.0319
## 4 1.2.20289.m1 133hpf 0.0207
## 5 1.2.21324.m1 133hpf 0.628
## 6 1.2.25539.m1 133hpf 1.27
## 7 1.2.1045.m1 29hpf -1.44
## 8 1.2.1655.m1 29hpf -0.204
## 9 1.2.20276.m1 29hpf -0.364
## 10 1.2.20289.m1 29hpf 0.0845
## 11 1.2.21324.m1 29hpf -0.681
## 12 1.2.25539.m1 29hpf -1.50
Question 10: Your answer should produce a tibble that looks like this;
## # A tibble: 6 × 2
## Stage hpf
## <chr> <dbl>
## 1 Egg 0
## 2 29hpf 29
## 3 133hpf 133
## 4 t2h 135
## 5 t12h 145
## 6 t72h 205
Question 11: Your answer should produce a tibble that looks like this;
## # A tibble: 36 × 5
## GeneID protein_name Stage Expression hpf
## <chr> <chr> <chr> <dbl> <dbl>
## 1 1.2.1045.m1 Homeobox protein NK-2 homolog E 133hpf 0.0328 133
## 2 1.2.1045.m1 Homeobox protein NK-2 homolog E 29hpf -1.44 29
## 3 1.2.1045.m1 Homeobox protein NK-2 homolog E Egg -1.67 0
## 4 1.2.1045.m1 Homeobox protein NK-2 homolog E t12h 2.55 145
## 5 1.2.1045.m1 Homeobox protein NK-2 homolog E t2h 0.259 135
## 6 1.2.1045.m1 Homeobox protein NK-2 homolog E t72h 1.27 205
## 7 1.2.1655.m1 Homeobox protein HLX1 133hpf 0.0221 133
## 8 1.2.1655.m1 Homeobox protein HLX1 29hpf -0.204 29
## 9 1.2.1655.m1 Homeobox protein HLX1 Egg -0.258 0
## 10 1.2.1655.m1 Homeobox protein HLX1 t12h 0.260 145
## # ℹ 26 more rows
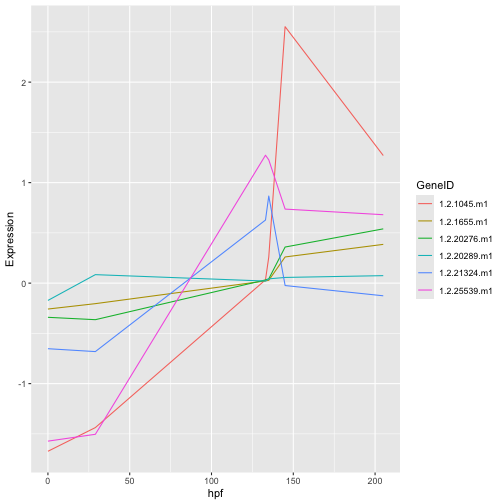
Question 12: Your answer should be a plot that looks like this;