BC3203
Expected outputs for Tutorial 6 exercises
Question 1: Your answer should be a function read_gff. If you run your function like this it should produce output as shown below
read_gff("cache/human_genes.gff")
## # A tibble: 42,220 × 9
## seqid source type start end score strand phase attributes
## <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr>
## 1 NC_000001.11 BestRefSeq gene 17369 17436 . - . ID=gene-MIR68…
## 2 NC_000001.11 Gnomon gene 29926 31295 . + . ID=gene-MIR13…
## 3 NC_000001.11 BestRefSeq gene 30366 30503 . + . ID=gene-MIR13…
## 4 NC_000001.11 BestRefSeq gene 34611 36081 . - . ID=gene-FAM13…
## 5 NC_000001.11 BestRefSeq gene 65419 71585 . + . ID=gene-OR4F5…
## 6 NC_000001.11 Gnomon gene 89006 267354 . - . ID=gene-LOC10…
## 7 NC_000001.11 BestRefSeq gene 134773 140566 . - . ID=gene-LOC72…
## 8 NC_000001.11 BestRefSeq gene 187891 187958 . - . ID=gene-MIR68…
## 9 NC_000001.11 Gnomon gene 200442 203983 . + . ID=gene-LOC10…
## 10 NC_000001.11 Gnomon gene 350706 476822 . - . ID=gene-LOC11…
## # ℹ 42,210 more rows
Question 2: Your answer should produce a tibble that looks like this;
## # A tibble: 42,220 × 10
## seqid source type start end score strand phase attributes length
## <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr> <dbl>
## 1 NC_000001.11 BestRe… gene 17369 17436 . - . ID=gene-M… 67
## 2 NC_000001.11 Gnomon gene 29926 31295 . + . ID=gene-M… 1369
## 3 NC_000001.11 BestRe… gene 30366 30503 . + . ID=gene-M… 137
## 4 NC_000001.11 BestRe… gene 34611 36081 . - . ID=gene-F… 1470
## 5 NC_000001.11 BestRe… gene 65419 71585 . + . ID=gene-O… 6166
## 6 NC_000001.11 Gnomon gene 89006 267354 . - . ID=gene-L… 178348
## 7 NC_000001.11 BestRe… gene 134773 140566 . - . ID=gene-L… 5793
## 8 NC_000001.11 BestRe… gene 187891 187958 . - . ID=gene-M… 67
## 9 NC_000001.11 Gnomon gene 200442 203983 . + . ID=gene-L… 3541
## 10 NC_000001.11 Gnomon gene 350706 476822 . - . ID=gene-L… 126116
## # ℹ 42,210 more rows
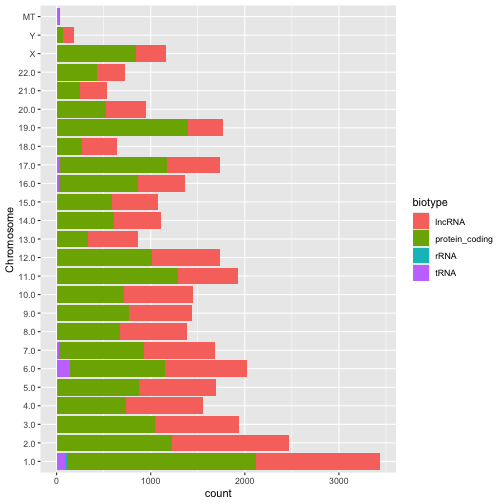
Question 3: Your answer should produce a plot that looks like this;
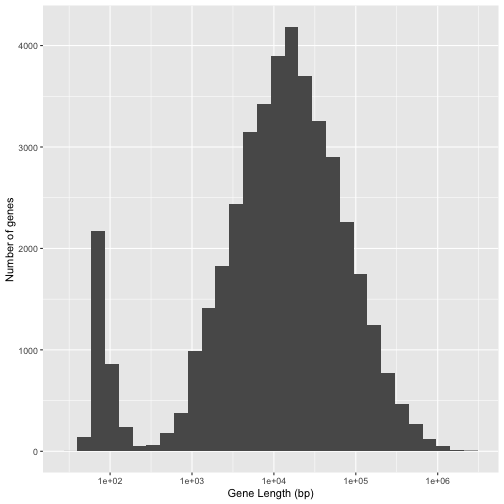
Question 4: Your answer should produce a plot that looks like this;
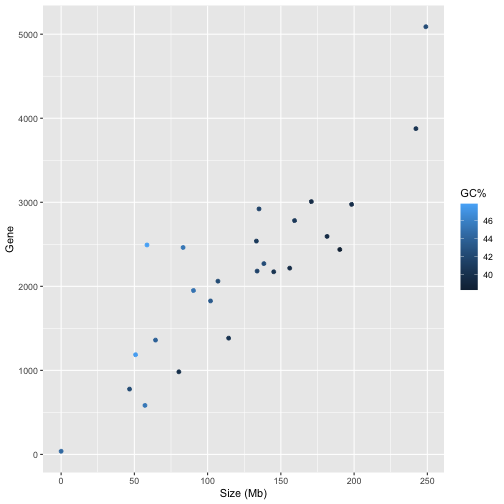
Question 5: Your answer should produce a tibble that looks like this;
## # A tibble: 37,478 × 20
## seqid source type start end score strand phase attributes Type Name
## <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 NC_0000… BestR… gene 17369 17436 . - . ID=gene-M… Chr 1.0
## 2 NC_0000… Gnomon gene 29926 31295 . + . ID=gene-M… Chr 1.0
## 3 NC_0000… BestR… gene 30366 30503 . + . ID=gene-M… Chr 1.0
## 4 NC_0000… BestR… gene 34611 36081 . - . ID=gene-F… Chr 1.0
## 5 NC_0000… BestR… gene 65419 71585 . + . ID=gene-O… Chr 1.0
## 6 NC_0000… Gnomon gene 89006 267354 . - . ID=gene-L… Chr 1.0
## 7 NC_0000… BestR… gene 134773 140566 . - . ID=gene-L… Chr 1.0
## 8 NC_0000… BestR… gene 187891 187958 . - . ID=gene-M… Chr 1.0
## 9 NC_0000… Gnomon gene 200442 203983 . + . ID=gene-L… Chr 1.0
## 10 NC_0000… Gnomon gene 350706 476822 . - . ID=gene-L… Chr 1.0
## # ℹ 37,468 more rows
## # ℹ 9 more variables: INSDC <chr>, `Size (Mb)` <dbl>, `GC%` <dbl>,
## # Protein <dbl>, rRNA <dbl>, tRNA <dbl>, `Other RNA` <dbl>, Gene <dbl>,
## # Pseudogene <dbl>
Question 6: Your answer should produce a tibble that looks like this;
## # A tibble: 25 × 2
## Name num_genes
## <chr> <int>
## 1 1.0 3673
## 2 2.0 2616
## 3 6.0 2115
## 4 11.0 2079
## 5 3.0 2061
## 6 19.0 1961
## 7 17.0 1883
## 8 12.0 1837
## 9 5.0 1797
## 10 7.0 1793
## # ℹ 15 more rows
Question 7: Your answer should produce a tibble that looks like this;
## # A tibble: 25 × 3
## # Groups: Name [25]
## Name chr_order num_genes
## <chr> <int> <int>
## 1 1.0 1 3673
## 2 2.0 2 2616
## 3 3.0 3 2061
## 4 4.0 4 1631
## 5 5.0 5 1797
## 6 6.0 6 2115
## 7 7.0 7 1793
## 8 8.0 8 1495
## 9 9.0 9 1542
## 10 10.0 10 1537
## # ℹ 15 more rows
Question 8: Your answer should produce a plot that looks like this;
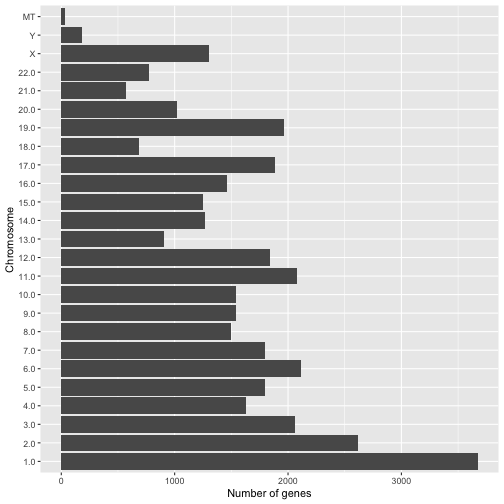
Question 9: Your answer should produce a plot that looks like this;
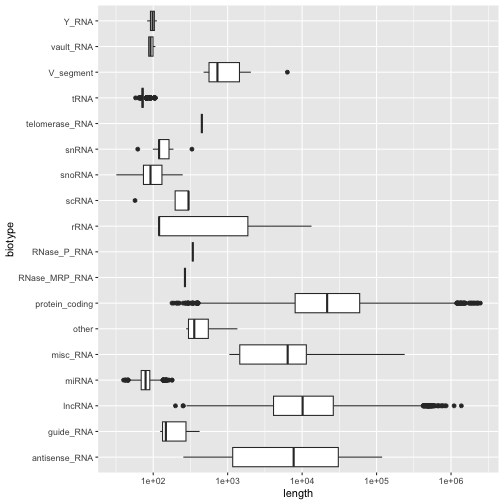
Question 10: Your answer should produce a plot that looks like this;